Contents
This script compares the CBlasso and the Blasso recovery in the case of a 3-spikes measure. The architecture is the following 1) We solve the SDP dual formulation of Blasso and CBlasso. For this step, 'cvx' solver is used. 2) We proceed to roots-finding for the previous computed dual polynomial. In this step, we identify a set of locations in which the support of the measure to reconstruct is included. 3) CBlasso : we alternate between Lasso steps and noise estimation 3bis) Blasso : we proceed to a Lasso step Some parts of this code heavily rely on the following works : (i) http://www.cims.nyu.edu/~cfgranda/scripts/superres_sdp.m (ii) http://nbviewer.jupyter.org/github/gpeyre/numerical-tours/blob/master/matlab/sparsity_8_sparsespikes_measures.ipynb
Developers : Claire Boyer, Yohann de Castro, Joseph Salmon (june 2016)clear all close all addpath(genpath('~/Documents/MATLAB/cvx')) ; addpath('math_tools/') ; rng(1); % Display features ms = 20; lw = 1; P = 2048*8; u = (0:P-1)'/P;Data generation
fc = 80; delta = 3/fc; % parameter for safe separation condition N = 2*fc+1; Fourier = @(fc,x)exp(-2i*pi*(-fc:fc)'*x(:)'); Phi = @(fc,x,a)Fourier(fc,x)*a; PhiS = @(fc,u,p)real( Fourier(fc,u)'* p ); % True signal x0 = [.5-delta .5 .5+2*delta]'; % spike positions a0 = [1 1 -1]'; % spike magnitudes n = length(x0); Phix0 = Phi(fc,x0,a0) ; % Noisy observations sigma = 1 ; w = randn(N,1) + 1i * randn(N,1); w = sigma * w; y = Phix0 + w; % Plot data f0 = PhiS(fc,u,Phix0); f = PhiS(fc,u,y); figure(1); hold on; set(gca,'FontSize',16) plot(u, [f0 f]); stem(x0, 10*sign(a0), 'k.--', 'MarkerSize', ms, 'LineWidth', 1); axis tight; box on; h=legend('${\mathcal{F}_n}^* \left( {\mathcal{F}_n} (\mu^0) \right)$', '${\mathcal{F}_n}^* y$'); set(h,'Interpreter','latex')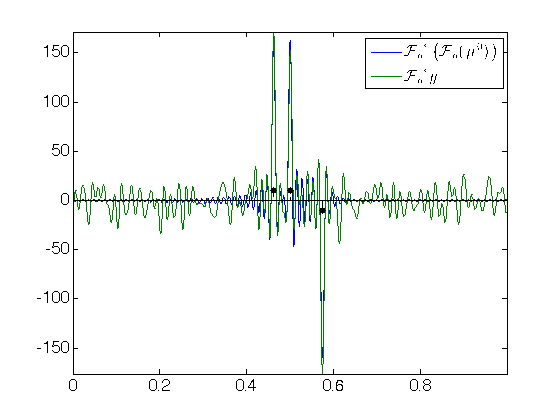
Blasso - constructing the dual polynomial
% Parameters for SDP resolution q = ones(N,1); alpha = 2 ; % SDP Blasso lambda_max_blasso = norm(PhiS(fc,u,y),inf) / N ; lambda_blasso = N * lambda_max_blasso/alpha; tic cvx_solver sdpt3 % SeDuMi % cvx_begin sdp quiet cvx_precision high; variable X(N+1,N+1) hermitian; variable c(N) complex; X >= 0; X(N+1,N+1) == 1; X(1:N,N+1) == c .* conj(q); trace(X) == 2; for j = 1:N-1, sum(diag(X,j)) == X(N+1-j,N+1); end if lambda_blasso==0 maximize(real( c' * y )) else minimize( norm(y/lambda_blasso-c, 'fro') ) end cvx_end Blasso_coeff = X(1:N,N+1); toc % Plot the Blasso polynomial etaLambda_Blasso = PhiS(fc,u,Blasso_coeff); figure(10) ; hold on; set(gca,'FontSize',16) ; stem(x0, sign(a0), 'k.--', 'MarkerSize', ms, 'LineWidth', lw); plot([0 1], [1 1], 'k--', 'LineWidth', lw); plot([0 1], -[1 1], 'k--', 'LineWidth', lw); plot(u, etaLambda_Blasso, 'b', 'LineWidth', lw); axis([0 1 -1.1 1.1]); set(gca, 'XTick', [], 'YTick', [0 1]); box on; title('Blasso')Elapsed time is 27.850022 seconds.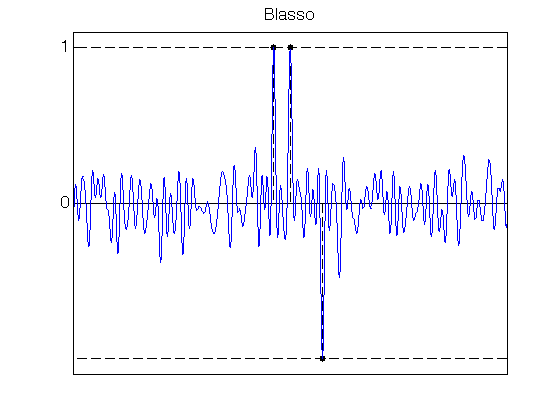
CBlasso - constructing the dual polynomial
% SDP CBLasso lambda_max_CBlasso = norm(PhiS(fc,u,y),inf) /(norm(y)*sqrt(N)) ; lambda_CBlasso = lambda_max_CBlasso/alpha ; tic cvx_precision best cvx_solver sdpt3 cvx_begin sdp quiet variable X(N+1,N+1) hermitian; variable c(N) complex; X >= 0; X(N+1,N+1) == 1; X(1:N,N+1) == c .* conj(q); trace(X) == 2; norm(c)<= 1/(sqrt(N)*lambda_CBlasso) ; for j = 1:N-1, sum(diag(X,j)) == X(N+1-j,N+1); end maximize(real(c'*y)) cvx_end toc CBlasso_coeff = X(1:N,N+1); % Plot the CBlasso polynomial etaLambda_CBlasso = PhiS(fc,u,CBlasso_coeff); figure(11) ; hold on; set(gca,'FontSize',16) ; stem(x0, sign(a0), 'k.--', 'MarkerSize', ms, 'LineWidth', lw); plot([0 1], [1 1], 'k--', 'LineWidth', lw); plot([0 1], -[1 1], 'k--', 'LineWidth', lw); plot(u, etaLambda_CBlasso, 'b', 'LineWidth', lw); axis([0 1 -1.1 1.1]); set(gca, 'XTick', [], 'YTick', [0 1]); box on; title('CBlasso')Elapsed time is 25.498846 seconds.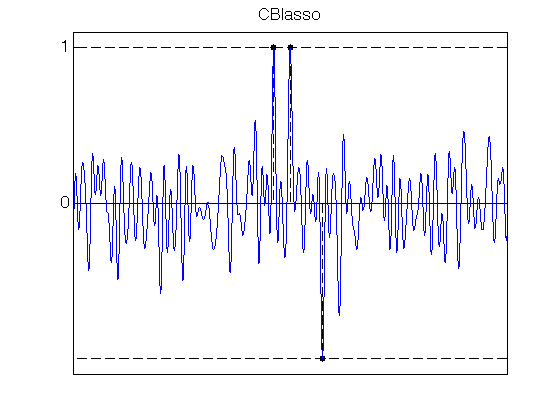
Roots-finding
tol = 1e-2 ; % CBlasso poly aux =- conv(CBlasso_coeff,flipud(conj(CBlasso_coeff))) ; % poly coeff of $- |p|^2$ aux(2*fc+1)=1+aux(2*fc+1); roots_all_CBlasso = roots(flipud(aux)); % roots function convention % CBlasso: Isolate roots on the unit circle roots_CBlasso_detected = roots_all_CBlasso(abs(1-abs(roots_all_CBlasso)) < tol); [~,I]=sort(angle(roots_CBlasso_detected)); roots_CBlasso_detected = roots_CBlasso_detected(I); % roots of 1-|p|^2 roots_CBlasso_detected = roots_CBlasso_detected(1:2:end); % roots of 1-|p % Blasso poly aux =- conv(Blasso_coeff,flipud(conj(Blasso_coeff))) ; % poly coeff of (- |p|^2) aux(2*fc+1)=1+aux(2*fc+1); roots_all_Blasso = roots(flipud(aux)); % roots function convention % Blasso: Isolate roots on the unit circle roots_Blasso_detected = roots_all_Blasso(abs(1-abs(roots_all_Blasso)) < tol); [~,I]=sort(angle(roots_Blasso_detected)); roots_Blasso_detected = roots_Blasso_detected(I); % roots of 1-|p|^2 roots_Blasso_detected = roots_Blasso_detected(1:2:end); % roots of 1-|p % Plot the complex roots figure(20) ; set(gca,'FontSize',16) plot(real(roots_all_CBlasso),imag(roots_all_CBlasso),'*'); hold on; plot(real(roots_CBlasso_detected),imag(roots_CBlasso_detected),'kp'); plot(cos(2*pi*x0), sin(2*pi*x0),'ro'); plot( exp(1i*linspace(0,2*pi,200)), '--' ); hold off; legend('Roots CBlasso','Roots detected','Support of x','Unit circle'); axis equal; axis([-1 1 -1 1]*1.3); figure(21) ; set(gca,'FontSize',16) plot(real(roots_all_Blasso),imag(roots_all_Blasso),'*'); hold on; plot(real(roots_Blasso_detected),imag(roots_Blasso_detected),'kp'); plot(cos(2*pi*x0), sin(2*pi*x0),'ro'); plot( exp(1i*linspace(0,2*pi,200)), '--' ); hold off; legend('Roots Blasso','Roots detected','Support of x','Unit circle'); axis equal; axis([-1 1 -1 1]*1.3);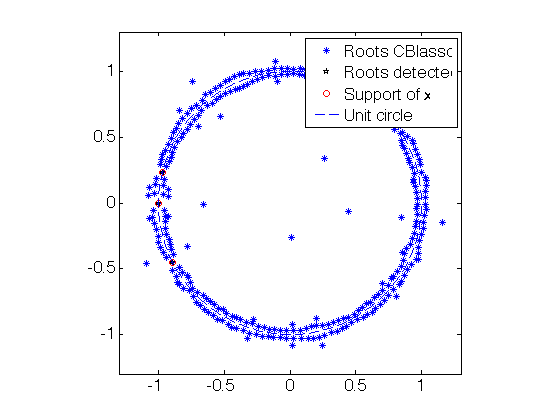
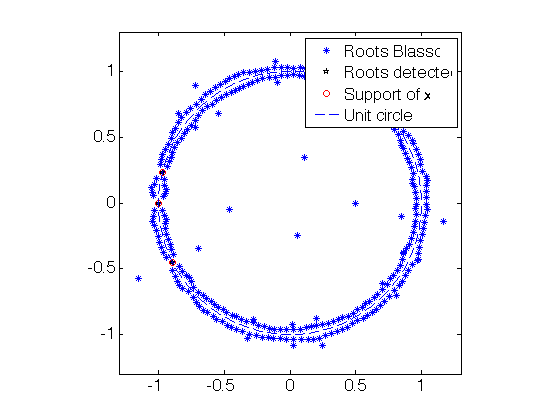
Solution via Blasso
% Precomputation for Blasso x_Blasso = angle(roots_Blasso_detected)/(2*pi); x_Blasso = sort(mod(x_Blasso,1)); Phix_Blasso = Fourier(fc,x_Blasso); sign_coeff_Blasso = sign(real(Phix_Blasso'*Blasso_coeff)); % Lasso step tic ; lasso_sol_1 = (Phix_Blasso\y - lambda_blasso*pinv(Phix_Blasso'*Phix_Blasso)*sign_coeff_Blasso ); toc ;Elapsed time is 0.002452 seconds.Solution via Scaled lasso (alternating Lasso step and noise estimation)
% Precomputation for CBlasso x_CBlasso = angle(roots_CBlasso_detected)/(2*pi); x_CBlasso = sort(mod(x_CBlasso,1)); Phix_CBlasso = Fourier(fc,x_CBlasso); sign_coeff_CBlasso = sign(real(Phix_CBlasso'*CBlasso_coeff)); % Alternating Lasso steps and noise estimations tol = 1e-4 ; maxiter = 1000 ; niter = 0 ; sigma_old = norm(y)/sqrt(N) ; sigma_hat = 0.1 ; % initialization for $\hat{\sigma}$ tic ; while (abs(sigma_hat(end) - sigma_old) > tol) && (niter<maxiter) niter = niter + 1 ; sigma_old = sigma_hat(end) ; lambda = sigma_hat(end) * lambda_CBlasso ; scaled_lasso_sol = (Phix_CBlasso\y - lambda*N*pinv(Phix_CBlasso'*Phix_CBlasso)*sign_coeff_CBlasso ); sigma_hat = norm(y - Phix_CBlasso * scaled_lasso_sol)/sqrt(2*N) ; % should divide by sqrt(2) end toc ;Elapsed time is 0.000915 seconds.Comparison plot : reconstructed measures via Blasso and CBlasso
figure(100); hold on; set(gca,'FontSize',16) stem(x0, a0, 'k.--', 'MarkerSize', ms, 'LineWidth', 1); stem(x_Blasso, lasso_sol_1, 'ro--', 'MarkerSize', ms/2, 'LineWidth', 1); stem(x_CBlasso, scaled_lasso_sol, 'b*--', 'MarkerSize', ms/2, 'LineWidth', 1); axis([0 1 -1.1 1.1]); legend('Original', 'BLasso','CBLasso');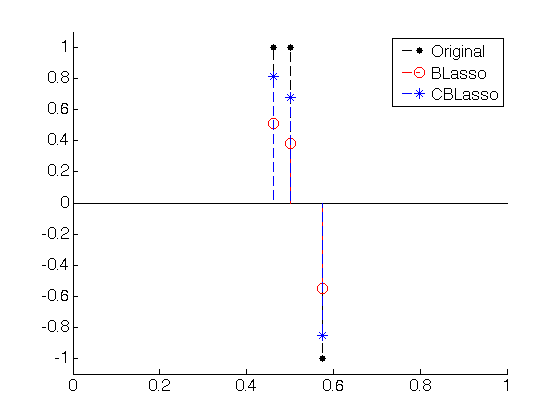
Noise estimation
sigma sigma_CBlasso = norm(y - Phix_CBlasso * scaled_lasso_sol)/sqrt(2*N) % divided by sqrt(2) due to the complex case sigma_Blasso = norm(y - Phix_Blasso * lasso_sol_1)/sqrt(2*N)sigma = 1 sigma_CBlasso = 0.9780 sigma_Blasso = 1.1467